The management of Black Sigatoka
Background: The Australian Banana Industry and Black Sigatoka
The value of the Australian banana industry is estimated to be AUD$350 million per annum. In 2001, nearly 238 000 tonnes of bananas were produced by 2100 growers across the nation. All Australian bananas are produced for consumption by local markets and ninety-five percent of bananas sold are of the Cavendish variety. The majority of bananas are grown in north Queensland, with 67% of the crop concentrated in the Tully, Cairns and Innisfail region.
In April 2001, the Australian banana industry suffered a potentially devastating outbreak of black Sigatoka in Tully, north Queensland. This was the first incursion in a major commercial growing region and failure to control the pathogen had the potential to have a significant impact on the industry as a whole.
The Queensland Department of Primary Industries (DPIQ) has had much success in eradicating previous incursions of black Sigatoka via plant destruction and replacement with resistant banana lines. Since the initial discovery of black Sigatoka in 1981 at Bamaga, an Aboriginal community located 40km from the tip of the Cape York Peninsula, the disease has been detected and eradicated in far north Queensland eight times. This ninth outbreak at Tully lies in north Queensland's major banana production area, where crops have an estimated value of AUD$119 million per annum (US$64 million) (Julianne Anderson, pers comm).
Management Response to the Tully Outbreak
 The following summary is based on a report titled: Peterson, R.A. (2002). Black-sigatoka eradication - controlled management program: Tully banana production area. Extensive surveys over the April-June period indicated the disease was restricted to the Tully valley area. More than 2700 samples were collected from all banana areas in north Queensland and laboratory tested, only 16 showed evidence of black Sigatoka.
The following summary is based on a report titled: Peterson, R.A. (2002). Black-sigatoka eradication - controlled management program: Tully banana production area. Extensive surveys over the April-June period indicated the disease was restricted to the Tully valley area. More than 2700 samples were collected from all banana areas in north Queensland and laboratory tested, only 16 showed evidence of black Sigatoka.
The low incidence of the organism over a limited area and lack of mature lesions in many samples indicated that the organism was only recently introduced to the area (1-2 wet seasons). In May/June 2001, less than 2 months after the detection of black Sigatoka at Tully, a plan to eradicate the disease was devised. The plan took advantage of the cool winter and dry spring conditions, during which the disease fails to develop. The eradication program involved:
- The removal of all diseased plant tissue from the plants and placing it on the ground to encourage decomposition. Black Sigatoka survival declines rapidly as the leaf tissue decomposes. If necessary this was repeated at 2-4 week intervals.
- Regulations were introduced to define the affected area (Tully Banana Protection Area - TBPA) and targets were set for the eradication program. Zero visible disease in the TPBA and 15% on any leaf in other areas.
- Where disease was detected growers were prevented from moving the fruit until all visible disease was removed.
- Regular inspections (4-6 week intervals) were carried out by trained, dedicated monitors, to ensure zero visible disease in the TBPA.
- An intense spray program (using both protectant and fungicide) was applied to prevent new infections and all banana plants outside plantations were de-leafed and the plant killed using herbicide injection.
- Sentinel plants were left unsprayed and monitored for disease.
- More than 8900 leaf samples were collected and tested for black Sigatoka between April and May 2002. All samples were laboratory tested for black Sigatoka.
- A program was put in place which would continue monitoring the TBPA and surrounding areas for 12 months from July 2002. The program included 6 rounds of intensive monitoring of all managed (plantation) banana plants, 12 rounds monitoring a 138 sites of unsprayed sentinel banana plants and 2 rounds of revisits to known sites of unmanaged banana plants.
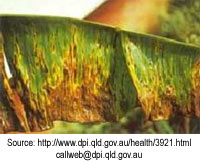 The results at the time the report was written suggest that black Sigatoka had been eradicated from the TBPA and that the target of 15% on any leaf outside the production area will be achieved. An important part of the success of the control of the black sigatoka outbreak at Tully was the ability to rapidly and accurately identify black Sigatoka on leaf samples. Read black Sigatoka diagnosis in Australia to find out how the diagnosis of black Sigatoka was improved.
The results at the time the report was written suggest that black Sigatoka had been eradicated from the TBPA and that the target of 15% on any leaf outside the production area will be achieved. An important part of the success of the control of the black sigatoka outbreak at Tully was the ability to rapidly and accurately identify black Sigatoka on leaf samples. Read black Sigatoka diagnosis in Australia to find out how the diagnosis of black Sigatoka was improved.
Black Sigatoka Diagnosis is Australia
Surveillance for Sigatoka leaf disease in banana is routinely carried out by DPIQ scientists at the Centre for Tropical Agriculture in Mareeba. Accurate diagnosis of black Sigatoka can be complicated by the morphological similarity of the related species Mycosphaerella musicola, the causal agent of yellow Sigatoka. In most cases, experienced plant pathologists can distinguish between yellow and black Sigatoka using symptom development and microscopic identification of fungal structures. However, where reproductive structures are absent due to prolonged rainfall, as happened during the recent Tully outbreak, morphological identification is not possible. In such cases, molecular methods are used to confirm diagnosis.
Strategies to improve Molecular Diagnosis
At the time of the outbreak the molecular methods available (sensu Johanson 1997) were slow and lacked specificity. The issue of specificity appeared to be due to the presence of geographically discrete populations of the fungus e.g. Papua New Guinea versus Pacific. The diagnostic test was suspected of being less powerful for the Torres Strait populations of the fungus. In July 2000, a joint project between DPIQ and the Cooperative Research Centre for Tropical Plant Protection (CRCTPP) was initiated to improve specificity of the procedure and increase sample processing speed in readiness for outbreaks of the disease. The project was co-managed by Dr Juliane Henderson (Research Officer, CRCTPP) and Mr Ron Peterson (Principal Plant Pathologist, Mareeba DPIQ). Components of the diagnostic test that required improvement included the way samples were collected and processed.
So, for example, the sampling technique was changed from cutting the lesions out of the banana leaves using a scalpel, to using a sterilised cork borer to reduce contamination. Other measures were to reduce cross contamination between samples, to improve the method for extracting DNA from the banana and to increase specificity and sensitivity of the test. The lab was also equipped to cope with more samples. The improvements lead to a definitive result in greater than 98% of samples. In addition, the improvements more than halved the extraction time and have increased daily throughput by more than eight-fold.
Application of the new molecular test
Transfer of the new technology in April 2001 coincided with Australia's most severe outbreak of black Sigatoka. This was the first outbreak in a commercial growing area; previous outbreaks have been much further north and in places where containment was much easier. Due to the high rainfall experienced in Tully, fungal structures were absent on banana samples collected for identification. As a result, diagnosis of up to 50% of samples had to be confirmed using the molecular test. This test provided Australian government bodies and the Australian banana industry the confidence to embark on a AUD$20 million surveillance and eradication plan in the Tully region and to date, over 2500 tests have been conducted.
Ensuring wuccess of the new eiagnostic assay
The CRCTPP together with DPIQ continues to monitor the use of the Sigatoka diagnostic test and researchers are exploring opportunities to automate and further improve the test. Organisations that rely heavily on accurate diagnostics will fund such studies . e.g. the Australian Banana Industry Protection Board (BIPB) and Horticulture Australia Limited (HAL). Some of the funded research will be directed towards genetic studies to track the source of the outbreak. Such a study may tell us how the 2001 Australian outbreak may have arisen (i.e. through single or multiple incursion).
Situation Update
Read (Henderson et al 2006) for an update of new molecular diagnostic technologies used to test for Black Sigatoka.
References
Hayden, H. 2001. Genetic variability in populations of pathogens causing black and yellow Sigatoka diseases of bananas. PhD Thesis, University of Queensland, Australia.
Johanson, A. 1997. Detection of Sigatoka Leaf Spot Pathogens of Banana by the Polymerase Chain Reaction. Natural Resources Institute, Chatham, United Kingdom.
Rychlik, W. 1993. Selection of primers for polymerase chain reaction. Pp 31-40 in Methods in Molecular Biology, Vol 15: PCR Protocols: Current Methods and Applications (B.A. White, ed.). Humana Press Inc, Totowa, New Jersey.
Stewart, C.N. & Via, L.E. 1993. A rapid CTAB DNA isolation technique useful for RAPD fingerprinting and other PCR applications. BioTechniques 14(5): 748-750.
White, T.J., Bruns, T., Lee, S. & Taylor, J.W. 1990. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. Pp 315-322 in PCR Protocols: A Guide to Methods and Applications (M.A. Innis, D.H. Gelfand, J.J. Sninsky and T.J. White, eds.). Academic Press Inc., San Diego, USA.
Henderson, J., Pattenmore, J. A., Porchun, S. C., Hayden, H. L., Van Brunschot, S., Grice, K. R. E., Peterson, R. A., Thomas-Hall, S. R. and Sitken, E. A. B. 2006. Black Sigatoka disease: new technologies to strengthen eradication strategies in Australia. Australasian Plant Pathology 35: 181-193.
Cost Benefit Analysis of Black Sigatoka Diagnostic
The improvements of the technique for laboratory based identification of black Sigatoka was subject to a cost-benefit analysis that conservatively indicates that every $8 spent on research has returned approximately $100 of savings to industry.
North Queensland is a major commercial banana-growing and the region around Tully, produces over $87 million worth of bananas, accounting for approximately 40% of Australian production.
The diagnostic was specifically developed to enable disease identification when spores are not available (e.g. after rain). The total research costs have been calculated at $480 980 (which includes costs of developing and implementing the test).
During the Tully outbreak, the diagnostic, designed by CRCTPP and DPIQ scientists, was used to confirm diagnosis of up to 50% of banana leaf samples entering the Mareeba laboratory, and played a major role in the surveillance strategy.
The diagnostic assay also provided government bodies and the banana industry the confidence to embark on a $20 million surveillance and eradication plan in the Tully region.
By February 2002, the Tully outbreak had cost the industry over $4 million in additional treatment costs (de-leafing and spraying) in an attempt to eradicate this disease. Use of the PCR-based black Sigatoka diagnostic contributed $140 800 to this bill, however early diagnosis of the disease also enabled rapid introduction of a management strategy.
This early warning actually reduced the number of de-leafing and spraying treatments, and their associated costs, that would have been necessary had there been a delay in identifying this outbreak. Most importantly, under the circumstances of this outbreak, without this test it is doubtful whether an eradication campaign would have been feasible at all, and black Sigatoka would probably have been declared endemic by now.
If black Sigatoka becomes established in the Tully Valley, a future potential loss of 25% from an endemic outbreak would cost the region almost $22 million in lost production and control costs, and additional de-leafing and spraying treatments would become routine. The effects of agricultural chemicals in sensitive environmental areas are also a concern, and impacts on the industry at the farm gate will have flow-on effects, even though these impacts have not been calculated as part of this cost-benefit analysis.
Clearly, the fact that the NADN was in the process of transferring the black Sigatoka diagnostic to the DPIQ Centre for Tropical Agriculture during this outbreak was a major factor in minimising the potential economic damage of this disease to the banana industry.